Digital Health Research Lab: In Depth Use Cases with EHRs (Wintersemester 2019/2020)
Dozent:
Prof. Dr. Erwin Böttinger
(Digital Health - Personalized Medicine)
,
Dr. Claudia Schurmann
(Digital Health - Personalized Medicine)
,
Jan-Philipp Sachs
(Digital Health - Personalized Medicine)
,
Ariane Morassi Sasso
(Digital Health - Personalized Medicine)
,
Suparno Datta
(Digital Health - Personalized Medicine)
,
PhD Riccardo Miotto
(Digital Health - Personalized Medicine)
,
Dr. Girish Nadkarni
(Digital Health - Personalized Medicine)
,
Benjamin Glicksberg
(Digital Health - Personalized Medicine)
,
Sedigheh Eslami
Allgemeine Information
- Semesterwochenstunden: 4
- ECTS: 6
- Benotet:
Ja
- Einschreibefrist: 01.10.-30.10.2019
- Lehrform: Seminar
- Belegungsart: Wahlpflichtmodul
- Lehrsprache: Englisch
- Maximale Teilnehmerzahl: 20
Studiengänge, Modulgruppen & Module
- SCAD: Scalable Computing and Algorithms for Digital Health
- HPI-SCAD-C Concepts and Methods
- SCAD: Scalable Computing and Algorithms for Digital Health
- HPI-SCAD-T Technologies and Tools
- SCAD: Scalable Computing and Algorithms for Digital Health
- HPI-SCAD-S Specialization
- DICR: Digitalization of Clinical and Research Processes
- HPI-DICR-C Concepts and Methods
- DICR: Digitalization of Clinical and Research Processes
- HPI-DICR-T Technologies and Tools
- DICR: Digitalization of Clinical and Research Processes
- HPI-DICR-S Specialization
- APAD: Acquisition, Processing and Analysis of Health Data
- HPI-APAD-C Concepts and Methods
- APAD: Acquisition, Processing and Analysis of Health Data
- HPI-APAD-T Technologies and Tools
- APAD: Acquisition, Processing and Analysis of Health Data
- HPI-APAD-S Specialization
- SAMT: Software Architecture & Modeling Technology
- HPI-SAMT-K Konzepte und Methoden
- SAMT: Software Architecture & Modeling Technology
- HPI-SAMT-T Techniken und Werkzeuge
- SAMT: Software Architecture & Modeling Technology
- HPI-SAMT-S Spezialisierung
- OSIS: Operating Systems & Information Systems Technology
- HPI-OSIS-K Konzepte und Methoden
- OSIS: Operating Systems & Information Systems Technology
- HPI-OSIS-T Techniken und Werkzeuge
- OSIS: Operating Systems & Information Systems Technology
- HPI-OSIS-S Spezialisierung
Beschreibung
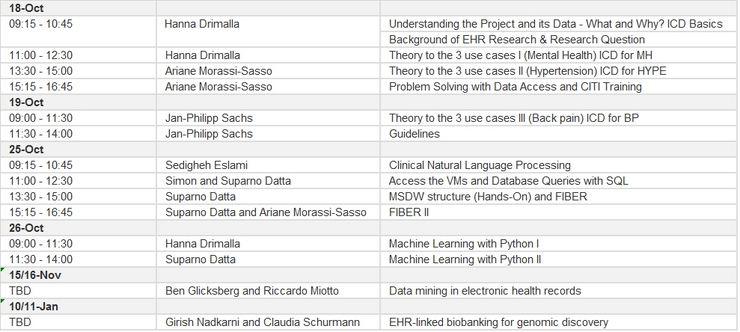
Note: The course 10/11 January will be postponed to 24/25 January!
The Digital Health Research Lab (DHRL) is a preparatory course for the master project of the research group of Professor Dr. Erwin Böttinger “Machine Learning on Real-World Health Data with Cloud-based In-Memory Database Computing” - but the course is also open to students not participating in this or another master project but interested in EHR and Mount Sinai Date Warehouse!
In the master project, students will apply machine learning methods using electronic health record (EHR) data of the Mount Sinai Data Warehouse (MSDW). To prepare you for this task, you will learn the basics about data access with FIBER (a python library for accessing MSHS Data Warehouse) and data analysis with Python by hands-on tasks on a small EHR data set.
The master project is structured around three use cases: heart disease, mental health, and back pain.
In the DHRL we will provide you with background knowledge about these topics, discuss current research questions in these fields and lay the foundation for the master project.
Last but not least, we will also teach you the fundamentals of clinical human research including ethical guidelines.
In Depth description for Block 3 (Nov. 15/16)
Background: Large-scale electronic health record (EHR) data have demonstrated the potential to completely transform the process of scientific discovery in precision medicine. Simply put, EHR data is any and all data that is collected during routine interactions with a hospital system, including clinical (e.g., diagnoses) and administrative (e.g., billing) information among many others. The ‘real world data’ contained within EHRs provide a tremendous amount of useful biomedical information that go beyond traditional experimental collection processes. Statistical and machine learning approaches applied to EHR data have led to important and clinically-relevant discoveries across many medical domains.
Problem: There are many roadblocks that come with working with EHR data, including infrastructure, quality control procedures, and addressing systemic and local biases. It is imperative that researchers interested in utilizing EHR data take into account the various limitations of the data in order to design effective and robust experiments.
Goal: The purpose of this course is to introduce students to the world of clinical informatics, with a particular emphasis on best practices for working with EHR data for high-impact projects. We will delve into the following topics:
- What data are contained in EHR?
- What are limitations to EHR data?
- What biases exist in such data and what are strategies to address them?
- How can other -omics data effectively be tied to EHR in an extensible mult-modal framework?
- What are common data models like OMOP and FHIR and why are they so important for EHR research?
- How to design a robust EHR-based studies and ask important questions?
- What are some state-of-the-art machine learning applications on EHR data?
- How can we move beyond manuscripts to translate findings from EHR data into the real world, such as the generation of real world evidence (RWE)?
This workshop will consist of lectures, interactive discussions, and simple exercises
In Depth description for Block 4 (Jan. 10/11)
"EHR-linked biobanking for genomic discovery" (Girish Nadkarni/Claudia Schurmann)
Precision medicine relies on the ability to assess disease risk at an individual level, detect early preclinical conditions and initiate preventive strategies using a combination of biological and clinical data. When treating diseases or trying to prevent them from developing, individual variability in genomic, environmental and lifestyle factors need to be taken into account. In this context, Electronic Health Records (EHRs) play an important role as they may enable better clinical decision making by integrating patient information from multiple sources.
In a combination of lectures and hands-on exercises you will learn about:
- The principles of clinical informatics while using EHR data to derive new phenotypes and get insight on how to use this data for genomic discovery.
- The Electronic Medical Records and Genomics (eMERGE) network and its contribution to genomics as poster child.
- Basics of genetic concepts, including inheritance, linkage disequilibrium, and types of genetic variation.
- Methodological questions related to large diverse data sets such as imputation and population stratification.
- Genome-wide association studies (GWAS) on different diseases for which all the phenotypic information was extracted from the EHR.
- Pleiotropy combined with phenome-wide association studies (PheWAS).
- Basics of polygenic risk scores and how they can be used.
- The use of genetic data together with EHR-derived clinical data in clinical settings.
Voraussetzungen
If you want to join the class, please contact asap Ariane Sasso for applying for access to the Mount Sinai Data Warehouse
Leistungserfassung
Grading will be based on four small group or individual projects or assignments (one per block), particularly in the form of reviewing important studies, writing "mini reviews", or hands-on exercises using the EHR data and presenting a summary of the results. The final grade will be determined by the individual parts whilst each part must be passed individually.
Termine
Note: The course 10/11 January will be postponed to 24/25 January!
Place G1 E 14/15
Zurück

