Bei einem Besuch in Südafrika haben die HPI-Professoren Lothar H. Wieler und Christoph Lippert unter anderem das Projekt "CHAMPS" kennengelernt.
>
Zum Artikel

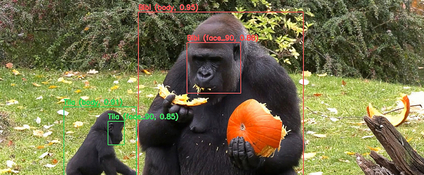
Mit dem "Gorilla Tracker" haben HPI-Forschende im Berliner Zoo ein Projekt getestet, das die Primatenforschung revolutionieren könnte.
>
Zum Artikel
Niclas Böhmer und Helene Kretzmer forschen und lehren künftig mit zwei Tenure-Track-Professuren am Hasso-Plattner-Institut.
>
Zum Artikel

Matthew Jörke und Lindsay Popowski von der Universität Stanford forschen am HPI an ihren Promotionsprojekten.
>
Zum Artikel


